Synthetic Biology
Research Lines
Using genetic engineering tools, we modify the genome of different bacteria for therapeutic purposes. We have different projects focused on different bacteria (Mycoplasma pneumoniae, Lactobacillus, Brucella) and different applications (lung cancer, glioblastoma, vaccines). Our group has a very powerful “translational” component by incorporating Pulmobiotics, a “start-up” that we founded in 2020 as a result of research conducted at the Center for Genomic Regulation.
1. Engineering Mycoplasma pneumoniae for the treatment of respiratory diseases (PULMOBIOTICS).
Using synthetic biology tools developed and patented, Pulmobiotics deleted virulent genes in M. pneumoniae, obtaining an attenuated strain known as MycoChassis. This MycoChassis is the base where new genes are added to design treatments for respiratory diseases.
Projects: PB_LC, MycoMatrix, PB_VAP, LactAS
2. Development of molecular engineering tools in other bacteria such as Lactobacillus and Brucella.
Projects: BruPatho, VENUS, Lac2Brain
BruPatho: Placental pathogenesis of brucella and development of new vaccines from a new molecular perspective in brucellosis
This project, funded in the PID2022 call "Proyectos de Generación de Conocimiento", aims to study the molecular mechanisms involved in Brucella pathogenesis using state-of-the-art systems biology technologies and apply the knowledge generated to develop vaccine candidates against B. suis. These studies are based on previous results from the group of our collaborator Professor Maria Jesús Grillo (IDAB-CSIC), which deal with the placental pathogenesis of B. melitensis and B. suis, as well as the spontaneous in vivo dissociation of B. suis bv2, together with the innovative molecular approaches that our group has successfully used in Mycoplasma pneumoniae.

VENUS: The vaginal microbiome as a model for the study of small proteins (SEPs)
Restoring microbiome dysbiosis by enriching with probiotic bacteria is currently used to treat infectious diseases. However, these treatments are often ineffective, especially in the presence of biofilms formed by pathogenic bacteria. Synthetic biology may pave the way for designing bacteria as “chassis” to eliminate biofilms. However, the impact of these chassis and their applicability to study microbiome homeostasis remains to be understood.
The Human Microbiome Project (HMB) allowed the massive identification of microbial genomes present in different human organs. These studies mainly annotated ORFs encoding large (>100aa) or smaller but conserved proteins. We found that, on average, 16% of the bacterial genome could encode small peptides (SEPs; <100aa), some of them essential and/or involved in infection and quorum sensing. Therefore, bacterial SEPs could be important for controlling the different human microbiomes, ensuring coexistence and preventing infections. However, identifying SEPs in microbial ecosystems and studying their roles in homeostasis is not straightforward.
In VENUS, we will address these challenges using the vaginal ecosystem due to its accessibility, available animal models and its relevance to health. First, we will use a software developed by us, RanSEPs, to identify secreted SEPs in the female reproductive tract (FRT). Second, after mimicking the vaginal ecosystem in vitro, we will experimentally characterize the function of the secreted SEPs and select those with antimicrobial or quorum sensing activity to rationally design different bacterial chassis based on bacteria found in the genital tract, with biofilm dispersal and homeostasis modulating activity. Finally, we will evaluate in vitro and in an in vivo mouse model the impact of the modified strains on microbiome homeostasis and biofilm degradation of S. aureus.
VENUS will address important concepts in Systems Biology by providing an understanding of the role of SEPs in the complex regulatory networks that orchestrate microbiome homeostasis and will use Synthetic Biology to translate this knowledge into the design of new therapeutic approaches. Project funded in the 2023 Call for Proposals “Europa Excelencia”.
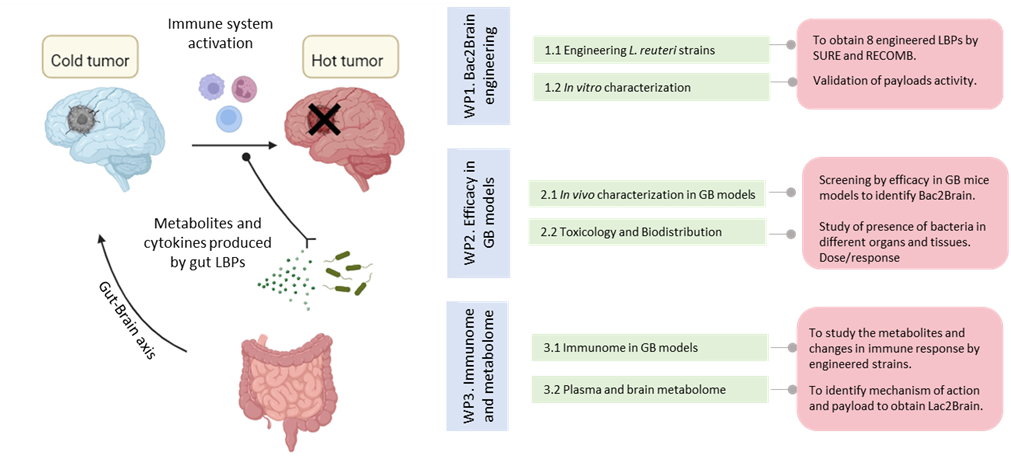
Lac2Brain: Engineering gut microbiota bacteria for glioblastoma treatment
Although immunotherapy has been transformative in the treatment of some cancers, brain malignancies show stagnant survival rates. In particular, glioblastoma (GB) is the most prevalent and aggressive primary brain tumor, with a median survival of 14 months.
The gut microbiome is a key modulator of health, including for neurological conditions. Indeed, there is a strong interconnection between the gut and the brain through a bidirectional network known as the gut-brain axis. Furthermore, there is growing evidence linking microbiome metabolism to the prognosis of different types of cancer, but none of these studies have examined the role of the microbiome in brain cancer. Finally, bacteria have a natural ability to colonize and proliferate within tumors.
We recently implemented a toolbox to modify the genome of a well-known probiotic strain of Lactobacillus that naturally inhabits the gastrointestinal tract. In this project, we intend to engineer this bacterium to secrete different bacterial metabolites and cytokines known to increase the immune response within the tumor microenvironment. After oral administration of the bacteria, these molecules will be secreted and translocated through the gut-brain axis to the brain, where they will exert their therapeutic effect, recovering the efficacy of immunotherapy.
In Lac2Brain, using "omics" approaches and in vivo mouse models, we will study the efficacy, mechanism of action and clinical potential of these treatments. The knowledge obtained in this proposal could be expanded in the future to treat different complex brain diseases. Project funded in the AECC LAB 2024 call.
Information of interest
MyLC: Mycoplasma pneumoniae as minimal lung bacterial chassis to deliver locally therapeutic proteins for the treatment of non-small cell lung carcinoma. (LC240083). Lluch Senar, M. (PI). FY24 Lung Cancer Research program (LCRP). Congressionally Directed Medical Research Programs (CDMRP). Department of Defence, USA. Execution: 2025-2027.
Patogenesis placentaria de brucella y desarrollo de vacunas desde una nueva Perspectiva molecular en brucelosis (BruPatho). (PID2022-139200OB-C22). Ministerio de Ciencia, Innovación e Universitat. Execution: 2023-2026.
El microbioma vaginal como modelo para el estudio de las proteínas pequeñas (SEPs) (EUR2023-143461). Ministerio de Ciencia e Innovación. Execution: 2022-2025.
Lac2Brain: Engineering bacteria from gut microbiota for the treatment of glioblastoma (LABAE247377LLUC). AECC. Convocatoria para consolidar grupos emergentes AECC lab. Execution: 2024-2027.
AYUDAS PARA CONTRATOS TORRES QUEVEDO 2020 : Estudio del potencial preclínico del primer bioterapéutico recombinante respiratorio para el tratamiento del enfisema pulmonar y la neumonía asociada a ventilación (PTQ2020-011049). Ministerio de Ciencia e Innovación. Execution: 2021-2024.
AYUDAS PARA CONTRATOS TORRES QUEVEDO 2020 : Ingeniería y desarrollo de un nuevo pulmobioterapéutico para el tratamiento del cáncer de pulmón (PTQ2020-011048). Ministerio de Ciencia e Innovación. Execution: 2021-2024. PULMOBIOTICS.
LactAs: desarrollo de un bioterapéutico vivo recombinante para el tratamiento del asma grave (CPP2021-008552: Proyecto de Colaboración Público-Privada Retos Colaboración). Ministerio de Ciencia e Innovación. Execution: 2022-2025. PULMOBIOTICS.
CLINICAL READINESS OF A LIVE BIOTHERAPEUTIC FOR TREATMENT OF NON-SMALL CELL LUNG CANCER (Grant agreement ID: 101098475). The European Innovation Council (EIC) HORIZON.3.1.Execution: 1 January 2023 a 30 June 2025. More info at Pulmobiotics. PULMOBIOTICS.
MycoMatrix: engineering of a recombinant live biotherapeutic to normalise tumour extracellular matrix in KRAS-driven Non-Small Cell Lung Cancer (CPP2022-009780: Proyecto de Colaboración Público-Privada Retos Colaboración). Ministerio de Ciencia, Innovación e Universitat. Execution: 2023-2026. PULMOBIOTICS.
NEOTEC 2021 (SNEO-20211019) - PLAN DE EMPRESA DE PULMOBIOTICS S.L. Centro para el Desarrollo Tecnológico Industrial (CDTI). Execution: 2022-2023. More info at Pulmobiotics. PULMOBIOTICS.
Broto A, Piñero-Lambea C, Segura-Morales C, Tio-Gillen AP, Unger WWJ, Burgos R, Mazzolini R, Miravet-Verde S, Jacobs BC, Casas J, Huizinga R, Lluch-Senar M, Serrano L. Engineering Mycoplasma pneumoniae to bypass the association with Guillain-Barré syndrome. Microbes Infect. 2024 Jul-Aug;26(5-6):105342. doi: 10.1016/j.micinf.2024.105342. Epub 2024 Apr 26. PMID: 38679229; PMCID: PMC11234194.
Miravet-Verde S, Mazzolini R, Segura-Morales C, Broto A, Lluch-Senar M, Serrano L. Author Correction: ProTInSeq: transposon insertion tracking by ultra-deep DNA sequencing to identify translated large and small ORFs. Nat Commun. 2024 Mar 27;15(1):2680. doi: 10.1038/s41467-024-47153-3. Erratum for: Nat Commun. 2024 Mar 7;15(1):2091. PMID: 38538589; PMCID: PMC10973512.
Lluch-Senar M. From science to business: translating live biotherapeutic products to the clinic. Nat Rev Bioeng. 2023 Jun 2:1-2. doi:10.1038/s44222-023-00078-w. Epub ahead of print. PMID: 37359770; PMCID:PMC10236387.
Mazzolini R, Rodríguez-Arce I, Fernández-Barat L, Piñero-Lambea C, Garrido V, Rebollada-Merino A, Motos A, Torres A, Grilló MJ, Serrano L, Lluch-Senar M. Engineered live bacteria suppress Pseudomonas aeruginosa infection in mouse lung and dissolve endotracheal-tube biofilms. Nat Biotechnol. 2023 Aug;41(8):1089-1098. doi: 10.1038/s41587-022-01584-9. Epub 2023 Jan 19. PMID: 36658340; PMCID: PMC10421741.
Burgos R, Garcia-Ramallo E, Shaw D, Lluch-Senar M, Serrano L. Development of a Serum-Free Medium To Aid Large-Scale Production of Mycoplasma-Based Therapies. Microbiol Spectr. 2023 Jun 15;11(3):e0485922. doi: 10.1128/spectrum.04859-22. Epub 2023 Apr 25. PMID: 37097155; PMCID: PMC10269708.
Montero-Blay A, Blanco JD, Rodriguez-Arce I, Lastrucci C, Piñero-Lambea C, Lluch-Senar M, Serrano L. Bacterial expression of a designed single-chain IL-10 prevents severe lung inflammation. Mol Syst Biol. 2023 Jan;19(1):e11037. doi: 10.15252/msb.202211037. Epub 2023 Jan 4. Erratum in: Mol Syst Biol. 2024 Mar;20(3):291-292. PMID: 36598022; PMCID: PMC9834763.
Piñero-Lambea C, Garcia-Ramallo E, Miravet-Verde S, Burgos R, Scarpa M, Serrano L, Lluch-Senar M. SURE editing: combining oligo-recombineering and programmable insertion/deletion of selection markers to efficiently edit the Mycoplasma pneumoniae genome. Nucleic Acids Res. 2022 Dec 9;50(22):e127. doi: 10.1093/nar/gkac836. PMID: 36215032; PMCID: PMC9825166.
Garrido V, Piñero-Lambea C, Rodriguez-Arce I, Paetzold B, Ferrar T, Weber M, Garcia-Ramallo E, Gallo C, Collantes M, Peñuelas I, Serrano L, Grilló MJ, Lluch-Senar M. Engineering a genome-reduced bacterium to eliminate Staphylococcus aureus biofilms in vivo. Mol Syst Biol. 2021 Oct;17(10):e10145. doi: 10.15252/msb.202010145. PMID: 34612607; PMCID: PMC8493563.
Shaw D, Miravet-Verde S, Piñero-Lambea C, Serrano L, Lluch-Senar M. LoxTnSeq: random transposon insertions combined with cre/lox recombination and counterselection to generate large random genome reductions. Microb Biotechnol. 2021 Nov;14(6):2403-2419. doi: 10.1111/1751-7915.13714. Epub 2020 Dec 16. PMID: 33325626; PMCID: PMC8601177.
Burgos R, Weber M, Martinez S, Lluch-Senar M, Serrano L. Protein quality control and regulated proteolysis in the genome-reduced organism Mycoplasma pneumoniae. Mol Syst Biol. 2020 Dec;16(12):e9530. doi: 10.15252/msb.20209530. PMID: 33320415; PMCID: PMC7737663.
Miravet-Verde S, Burgos R, Delgado J, Lluch-Senar M, Serrano L. FASTQINS and ANUBIS: two bioinformatic tools to explore facts and artifacts in transposon sequencing and essentiality studies. Nucleic Acids Res. 2020 Sep 25;48(17):e102. doi: 10.1093/nar/gkaa679. PMID: 32813015; PMCID: PMC7515713.
Piñero-Lambea C, Garcia-Ramallo E, Martinez S, Delgado J, Serrano L, Lluch-Senar M. <i>Mycoplasma pneumoniae</i> Genome Editing Based on Oligo Recombineering and Cas9-Mediated Counterselection. ACS Synth Biol. 2020 Jul 17;9(7):1693-1704. doi: 10.1021/acssynbio.0c00022. Epub 2020 Jun 22. PMID: 32502342; PMCID: PMC7372593.
Montero-Blay A, Miravet-Verde S, Lluch-Senar M, Piñero-Lambea C, Serrano L. SynMyco transposon: engineering transposon vectors for efficient transformation of minimal genomes. DNA Res. 2019 Aug 1;26(4):327-339. doi: 10.1093/dnares/dsz012. PMID: 31257417; PMCID: PMC6704405.
Miravet-Verde S, Ferrar T, Espadas-García G, Mazzolini R, Gharrab A, Sabido E, Serrano L, Lluch-Senar M. Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol. 2019 Feb 22;15(2):e8290. doi: 10.15252/msb.20188290. PMID: 30796087; PMCID: PMC6385055.
Lluch-Senar M, Delgado J, Chen WH, Lloréns-Rico V, O'Reilly FJ, Wodke JA, Unal EB, Yus E, Martínez S, Nichols RJ, Ferrar T, Vivancos A, Schmeisky A, Stülke J, van Noort V, Gavin AC, Bork P, Serrano L. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol Syst Biol. 2015 Jan 21;11(1):780. doi: 10.15252/msb.20145558. PMID: 25609650; PMCID: PMC4332154.
Funding
Additional information