Bioinformatics of Genome Diversity
Research Lines
The Genomic Diversity Bioinformatics group aims to explain the nature of genome variation and its relationship to phenotypic variation and fitness. A new dimension in genetic variation studies is provided by the complete genomes that are increasingly being sequenced (e.g., Project 1000 Genomes ), as well as new high-throughput data from other omic layers, such as the transcriptomic and epigenomic data of the ENCyclopedia Of Dna. Elements (ENCODE/modENCODE) or the International Human Epigenome Consortium (IHEC). We follow an interdisciplinary approach, fusing methods and knowledge from genomics, population genetics, data science, systems biology and bioinformatics, to address, both in Drosophila and in humans, the different planned objectives.
We currently have the following lines of research:
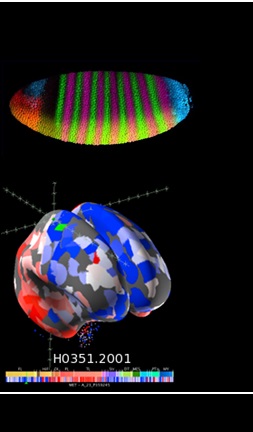
Mapping selection on embryo development in Drosophila and human brain
Integrate information from population genomics data with recent knowledge on developmental genetics (Drosophila), single-cell genomics and brain transcriptomics (humans) to obtain evidence for the evolution of embryonic development and human brain evolution at the microevolutionary level and test the selective importance of hierarchy in gene networks, pleiotropic interactions or preferential evolution of regulatory vs. coding sequences.
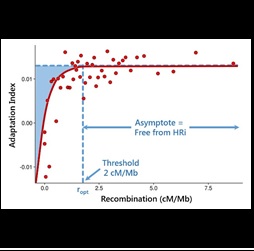
Define parameters that measure the adaptive potential of a genome
Recent evidence based on patterns of genome diversity shows that many loci and regions of the genome are affected by recurrent linked selection (genetic draft). Our goal is to develop a new general statistical approach to estimate the fraction of adaptive substitutions for any region of the genome, regardless of whether or not they are experiencing recurrent linked selective events.

PopFly and PopHuman, the reference population genomics browsers for Drosophila and humans
Update your browsers with new genomic data generated after the release of the PopFly and PopHuman genome browsers and complement them with data from other species to make them the reference population genomics browsers for eukaryotes.
Information of interest
Funded projects
Scientific
2018-2020: Genòmica poblacional de ladaptació. Researchers: Mauro Santos (PI1) and Antonio Barbadilla (PI2), Alfredo Ruiz, Antoni Fontdevila, Pilar Garcia-Guerreiro, Francisco Rodríguez-Trelles, Sònia Casillas, Raquel Egea, Marta Coronat, Sergi Hervàs, Jesús Murga. MINECO CGL2017-89160-P. 2018-2020 (In execution).
2018-2020: Genòmica, Bioinformàtica i Biologia Evolutiva. Grup recerca consolidat de la Generalitat de Catalunya ( 2017 SGR 1379. Alfredo Ruiz (PI), Mauro Santos, Antonio Barbadilla, Mario Càceres, Isaac Salazar. 2018-2020 (In execution).
2014-2017: La interpretació de la variació genòmica des de la seqüència nucleotídica al fenotip a Drosophila i humans. Investigadors: Antonio Barbadilla (IP1), Mario Càceres (IP2), Isaac Salazar, Sònia Casillas, Marta Puig, Raquel Egea, Carla Giner, Marta Coronada, Sergi Hervàs, Isaac Noguera, Ion Lerga. MINECO BFU2013-42649-P. 2014-2017 (Completed)
2014-2016: Genòmica, Bioinformàtica i Biologia Evolutiva. Grup de recerca consolidat de la Generalitat de Catalunya ( 2014 SGR 1346). Alfredo Ruiz (PI), Mauro Santos, Antonio Barbadilla, Mario Cáceres, Isaac Salazar. 2014-2016. (Completed)
2010-2013: Anàlisi genòmica del polimorfisme i la divergència a Drosophila melanogaster . Investigadors: Antonio Barbadilla (PI), Sònia Casillas, Esther Betrán, David Castellà, Miquel Ràmia, Maite Barrón. MICINN BFU2009-09504/BMC. 2010-2013. (Completed)
2008-2012: Panell de referència genètica de Drosophila: un recurs comunitari per a l'estudi de la variació genotípica i fenotípica. PIs: Trudy Mackay, Stephen Richards i Richard Gibbs (Completed) http://flybase.org/static_pages/news/whitepapers/Drosophila_Genetic Reference Panel_Whitepaper.pdf
2009–2013: Genòmica, bioinformàtica i evolució. Grup de recerca consolidat de la Generalitat de Catalunya ( 2009 SGR 88). Alfredo Ruiz (PI), Antonio Barbadilla, Roger Vila i Isaac Salazar. 2009-2013. (Completed)
2007-2009: Patterns de nucleotide diversity in diferents functional regions de Drosophila and Chordates. Investigadors: Antonio Barbadilla (PI), Sònia Casillas, Natalia Petit, Raquel Egea, Esther Betrán. MEC BFU2006-08640/BMC. (Completed)
2005-2008: Estudis associatius assistits per Inferència i Tecnologies Semàntiques (ASSIST). Coordinador del projecte: P. Mitkas (CERHT/ITI). Sisè Programa Marc. STRP: Informació biomèdica integrada per a una millor salut. FP6-IST-2004-027510. PI UAB: Antonio Barbadilla (Completed)
2005: Identificació de gens implicats en la susceptibilitat de patir fibromiàlgia i/o síndrome de fatiga crònica. Similituds i diferències genètiques. Fundació per a la Fibromiàlgia i la Síndrome de Fatiga Crònica, Fundació Echevarne, ebiointel, Dpt Genètica i Microbiologia UAB, CEGEN, banc ADN. (Ended)
2003-2005: Genòmica comparada i funcional de Drosophila: causes i conseqüències de les reordenacions cromosòmiques naturals. Investigadors: A. Ruiz (PI), A. Barbadilla, Josefa González, Ferran Casal, Bàrbara Negre, Marta Puig, Cristina Santa, Sònia Casillas . MCYT - BMC2002-01708. 2003-2005. (Completed)
1999-2002: Projecte: Distribució de les taxes de recombinació local causades per conversió i entrecreuament al llarg del genoma. Investigadors: A. Barbadilla (PI); A. Navarro, Cristina Santa. DGICYT - PB98-0900-C02-02. 1999-2002. (Completed)
1996-1998: Origen i evolució molecular de les inversions cromosòmiques de Drosophila buzzatii. Investigadors: A. Ruiz (PI); A. Barbadilla; E. Betrán; A. Navarro; José María Ranz; Mario Càceres; Josefa González. DGICYT-PB95-0607. (Completed)
1994-1996: Significat adaptatiu del polimorfisme cromosòmic. Investigadors: A. Ruiz (PI); A. Barbadilla; E. Betrán; A. Navarro. DGICYT – PB93-0844. (Completed)
Technological
2006-2012: Projecte Plataforma Bioinformàtica UAB-Instituts UAB-Instituts de Recerca Hospitalària / Plataforma Bioinformàtica UAB-Instituts de Recerca Hospitalària . A. Barbadilla (PI), S. Casillas, Àlex Sànchez, José Manuel Soria. Vicerectorat de Projectes Estratègics UAB, 2006-12. (Completed)
2004-2005: Projecte Neotec/La Caixa per a l'execució del projecte ebioPlatform : / Neotec /La Caixa project for the developing of ebioPlatform . I. Durán, A. Barbadilla, B. Aranda, J. Puiggene, J. Colomer, N. Rierol. Ministeri dIndústria, 2004-05. (Completed)
2002: Gestió del desenvolupament d'eines bioinformàtiques / Management for the development of bioinformatic tools . PI: A. Barbadilla. Conveni col·laboració Fundació Empresa i Ciència, UAB, 2002. (Completed)
2001-2002: Projecte del Centre d'Innovació i Desenvolupament Empresarial (CIDEM) per a la creació d'una empresa de base tecnològica / CIDEM projecte per desenvolupar technological based company. PI: A. Barbadilla. 2001-02. (Completed)
Communicating Science
Genètica i Genòmica: una invitació a la genètica. Projecte d'accions especials de difusió de la ciència i la tecnologia. Ministeri Ciència i Tecnologia. / Special action project for public communication of science. Researchers: A. Barbadilla (PI); C. Santa. AE00-0128-DIF. 2001-02. (Completed)
Una invitación a la bioinformática. Any de la Computació. UAB 2008. PI: Antonio Barbadilla. (Completed)
Adapnation: melanogaster catch the fly . Sònia Casillas (PI), Robert Torres, Josefa González, Antonio Barbadilla. 2019-2020. Fundació espanyola per a la ciència i la tecnologia. FCT-19-14835. (Completed)
Teaching
2019-2021 Adapta't . Teaching quality improvement project subsidized by the Vice-Rectorate for Academic Programming and Quality of the UAB. Professors: Sònia Casillas (PI), Gemma Armengol, Antonio Barbadilla, Aina Colomer, Marta Coronat, Raquel Egea, Pilar Garcia, Ruth Gómez, Jesús Murga, Alicia Roque. (Completed)
2011-2012: Coordinator Proposal and implementation of the UAB Strategic Master's in Bioinformatics. Strategic action of the UAB-Campus of Excellence project. (2011-2012). (Completed)
2007-2009: Coordinator Proposal and implementation of the UAB Genetics Degree. Faculty of Biosciences. (2007-2009). (Completed)
2002-2019: Design and implementation of a Web 2.0 platform for university teaching (annual update). Genomics and Bioinformatics Service (2002-2019)
2007-2008: Permanent Genetics Classroom. Teaching Innovation Project Grants 2007. Vice-Rectorate for Studies and Quality, UAB. 2007-2008. Antonio Barbadilla (PI), Miquel Ràmia, Maite Barrón, Santiago Pastor, Ricard Marcos, Amadeu Creus, Antonia Velázquez. (Completed)
2004-2006: Implementation of multimedia tools in an entire Degree: the case of Biology at the UAB / Implementation of multimedia tools at the University of College: the case of Biology at the UAB. (Together with the Biology teaching committee: Javier Retana, Amalia Molinero, Marina Luquín, Miquel Ninyerola and Elena Ibáñez). Projects to improve teaching quality AGAUR 2004MQD 00068. 2004-06. (Completed)
Rodriguez-Galindo, M., Casillas, S., Weghorn, D. and Barbadilla, A. 2020. Germline de novo mutation rates on exons versus introns in humans. Nat Commun 11, 3304 (2020). https://doi.org/10.1038/s41467-020-17162-z
Casillas*, S., R. Mulet*, P. Villegas-Mirón, S. Hervás, E. Sanz, D. Velasco, J. Bertranpetit, H. Laayouni & A. Barbadilla. 2018. PopHuman: the human population genomics browser. Nucl. Acids Res. gkx943, https://doi.org/10.1093/nar/gkx943
Salvador*, I., M. Coronado*, D. Castellano, A. Barbadilla & I. Salazar. 2018. Mapping selection within Drosophila melanogaster embryo’s anatomy. Molecular Biology and Evolution https://doi.org/10.1093/molbev/msx266.
Casillas, S. and A. Barbadilla. 2017. Molecular Population Genetics. Genetics 205: 1003–1035. https://doi.org/10.1534/genetics.116.196493
Huang, W. et al. 2014. Natural Variation in Genome Architecture. Among 205 Drosophila melanogaster Genetic Reference Panel Lines. Genome Research 2014. 24:1193-120.
T.F.C. Mackay*, S. Richards*, E.A. Stone*, A. Barbadilla*, ..., S. Casillas, M. Barrón, D. Castellano, P. Librado, M. Ràmia, J. Rozas et al. 2012. The Drosophila melanogaster Genetic Reference Panel: A Community Resource for Analysis of Population Genomics and Quantitative Traits. Nature 482: 173-178. * Equal contribution.
Egea, R., S. Casillas and A. Barbadilla (2008). Standard & Generalized McDonald and Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 2008 July 1; 36 (Web Server issue): W157–W162.
Cáceres, M., J.M. Ranz, A. Barbadilla, M. Long & A. Ruiz. (1999). Generation of a widespread Drosophila inversion by a transposable element, Science 285: 415-418.
All publications
Articles a International Science Journals (JCR)
Kapun, M. et al. 2021. Drosophila Evolution over Space and Time (DEST) - A New Population Genomics Resource. bioRxiv 2021.02.01.428994; doi: https://doi.org/10.1101/2021.02.01.428994
Rodriguez-Galindo, M., Casillas, S., Weghorn, D. and Barbadilla, A. 2020. Germline de novo mutation rates on exons versus introns in humans. Nat Commun 11, 3304 (2020). https://doi.org/10.1038/s41467-020-17162-z
Murga-Moreno, J.; Coronado-Zamora, M.; Hervás, S.; Casillas, S & A. Barbadilla 2019. iMKT: the integrative McDonald and Kreitman test. Nucleic Acids Research https://doi.org/10.1093/nar/gkz372
Coronado*, M, I. Salvador*, D. Castellano, A. Barbadilla & I. Salazar. 2019. Adaptation and conservation throughout the Drosophila melanogaster life-cycle. Genome, Biology and Evolution evz086, https://doi.org/10.1093/gbe/evz086.
Murga-Moreno, J.; Coronado-Zamora, M.; Bodelón, A.; Barbadilla, A. and Casillas, S 2019. PopHumanScan: the online catalog of human genome adaptation. Nucleic Acids Research, gky959, https://doi.org/10.1093/nar/gky959
Casillas*, S., R. Mulet*, P. Villegas-Mirón, S. Hervás, E. Sanz, D. Velasco, J. Bertranpetit, H. Laayouni & A. Barbadilla. 2018. PopHuman: the human population genomics browser. Nucl. Acids Res. gkx943, https://doi.org/10.1093/nar/gkx943 *Equal contribution.
Salvador*, I., M. Coronado*, D. Castellano, A. Barbadilla & I. Salazar. 2018. Mapping selection within Drosophila melanogaster embryo’s anatomy. Molecular Biology and Evolution https://doi.org/10.1093/molbev/msx266.
* Equal contribution.
S. Hervas, E. Sanz, S. Casillas, J. Pool and A. Barbadilla. 2017. PopFly: the Drosophila population genomics browser. Bioinformatics https://doi.org/10.1093/bioinformatics/btx301
Casillas, S. and A. Barbadilla. 2017. Molecular Population Genetics. Genetics 205: 1003–1035. https://doi.org/10.1534/genetics.116.196493
Pavlidou, P.; A. Daponte, R. Egea, E. Dardiotis, G.M. Hadjigeorgiou, A. Barbadilla & T. Agorastos. 2016. Genetic polymorphisms of FAS and EVER genes in a Greek population and their susceptibility to cervical cancer: a case control study. BMC Cancer 2016 16:923.
Castellano, D., M. Coronado-Zamora, JL. Campos, A. Barbadilla, A. Eyre-Walker. 2016. Adaptive evolution is substantially impeded by Hill-Robertson interference in Drosophila. Mol Biol Evol (2016) 33 (2): 442-455.
Guillén, Y. et al. 2015. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol Evol (2015) 7 (1):349-366.doi: 10.1093/gbe/evu291.
Huang, W. et al. 2014. Natural Variation in Genome Architecture. Among 205 Drosophila melanogaster Genetic Reference Panel Lines. Genome Research 2014. 24:1193-120.
A. Martínez-Fundichely, S. Casillas, R. Egea, M. Ràmia, A. Barbadilla, L. Pantano, M. Puig & M. Cáceres. 2014. InvFEST, a database integrating information of polymorphic inversions in the human genome. Nucl. Acids Res.42 (D1): D1027-D1032.doi: 10.1093/nar/gkt1122.
Gutiérrez F,M Gárriz,JM. Peri, L Ferraz,D Sol, JB Navarro, A Barbadilla, M Valdés. 2013. Fitness costs and benefits of personality disorder traits. Evolution and Human BehaviorFitness costs and benefits of personality disorder traits. Evolution and Human Behavior. Volume 34, Issue 1, January 2013, Pages 41–48.
T.F.C. Mackay*, S. Richards*, E.A. Stone*, A. Barbadilla*, ..., S. Casillas, M. Barrón, D. Castellano, P. Librado, M. Ràmia, J. Rozas et al. 2012. The Drosophila melanogaster Genetic Reference Panel: A Community Resource for Analysis of Population Genomics and Quantitative Traits. Nature 482: 173-178.
* Equal contribution.
Ràmia M, Librado P, Casillas S, Rozas, J & Barbadilla A. 2012. PopDrowser: The Population Drosophila Browser. Bioinformatics Feb 15;28(4):595-6.
Miquel M, López-Ribera I, Ràmia M, Casillas S, Barbadilla A, Vicient CM. 2011. MASISH: a database for gene expression in maize seeds. Bioinformatics 27(3):435-436
Petit, N. and A. Barbadilla (2009). The efficiency of purifying selection in Mammals vs Drosophila metabolic genes. J. Evol. Biology 22: 2118-2124.
Petit, N. and A. Barbadilla (2009). Selection efficiency and effective population size in Drosophila species. J. Evol. Biology 22: 515-526.
Egea, R., S. Casillas and A. Barbadilla (2008). Standard & Generalized McDonald and Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 2008 July 1; 36 (Web Server issue): W157–W162.
Casillas, S., R. Egea, N. Petit, C. Bergman and A. Barbadilla (2007). Drosophila Polymorhism Database (DPDB): a portal for nucleotide polymorphism in Drosophila. Fly 1 (4): 205-211.
Casillas, S., A. Barbadilla and C. Bergman (2007). Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol doi:10.1093/molbev/msm150.
Petit, N., S Casillas, A. Ruiz and A. Barbadilla (2007). Protein polymorphism is negatively correlated with conservation of intronic sequences and complexity of expression patterns in Drosophila melanogaster. Journal of Molecular Evolution 64: 511-518.
Egea, R, S. Casillas, E. Fernández, MA Senar & A. Barbadilla (2007). MamPol: a database of nucleotide polymorphism in the Mammalia class. Nucleic Acids Res. 2007 January; 35(Database issue): D624–D629
Casillas, S, B Negre, A Barbadilla and A Ruiz (2006). Fast sequence evolution of Hox and Hox-derived genes in the genus Drosophila. BMC Evolutionary Biology 2006, 6:106.
Casillas, S & A. Barbadilla (2006). PDA v.2: improving the exploration and estimation of nucleotide polymorphism in large data sets of heterogeneous DNA. Nucl. Acids Res. 2006 34: W632-W634.
Casillas, S, N. Petit & A. Barbadilla (2005). DPDB: a database for the storage, representation and analysis of polymorphism in the Drosophila genus. Bioinformatics 2005 21: ii26-ii30; doi:10.1093/bioinformatics/bti1103
Bárbara Negre, Sònia Casillas, Magali Suzanne, Ernesto Sánchez-Herrero, Michael Akam, Michael Nefedov, Antonio Barbadilla, Pieter de Jong and Alfredo Ruiz (2005). Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Research 15: 692-700, 2005
Casillas, S & A. Barbadilla (2004). PDA: a pipeline to explore and estimate polymorphism in large DNA databases. Nucleic Acid Research, Web Server issue 32: W166-W169.
Casillas, S., N Petit, R Egea, A Barbadilla. 2004. Storage, Representation and Analysis of Nucleotide Polymorphism. Spanish Bioinformatics Conference, 137-141.
Navarro, A. Barbadilla & A. Ruiz. (2000). Effect of inversion polymorphism on the neutral nucleotide variability of linked chromosomal regions. Genetics 155: 685-698.
Cáceres, M., J.M. Ranz, A. Barbadilla, M. Long & A. Ruiz. (1999). Generation of a widespread Drosophila inversion by a transposable element, Science 285: 415-418.
Cáceres, M., A. Barbadilla & A. Ruiz. (1999). Recombination rate predicts inversion size in Diptera», Genetics 153: 251-259.
Cáceres, M., A. Barbadilla & A. Ruiz. (1997). Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: Is inversion length a selected trait?, Evolution 51: 1149-1155.
Betrán, E., J. Rozas, A. Navarro & A. Barbadilla. (1997). The estimation of the number and the distribution of gene conversion tracts from population DNA data», Genetics 146: 89-99.
Navarro, A., E. Betrán, A. Barbadilla & A. Ruiz. (1997). Recombination and gene flux caused by crossing over and gene conversion in inversion heterokaryotypes. Genetics 145: 281-295.
Ruiz A., J.M., Ranz, M. Caceres, C. Segarra, A. Navarro & A. Barbadilla. (1997). Chromosomal evolution and comparative gene mapping in the Drosophila repleta species group. Genetics and Mol. Biol. 20: 553-565.
Barbadilla A, King LM, Lewontin RC. What does electrophoretic variation tell us about protein variation? Mol Biol Evol. 1996 Feb;13(2):427-32
Ruiz, A. y A. Barbadilla. (1995). The contribution of quantitative trait loci and neutral marker loci to the genetic variances and covariances among quantitative traits in random mating populations», Genetics 139: 445-455.
Barbadilla, A. Ruiz, M. Santos & A. Fontdevila. (1994). Mating pattern and fitness component analysis associated with inversion polymorphism in a natural population of Drosophila buzzatii, Evolution 48: 767-780.
Naveira, H, & A. Barbadilla. (1992). The theoretical distribution of lengths of intact chromosome segments around a locus held heterozygous with backcrossing in a diploid species, Genetics 130: 205-209.
Barbadilla, A., H. Naveira, A. Ruiz & M. Santos. (1992). The estimation of genotypic probabilities in an adult population by the analysis of descendants, Genetical Research 59: 131/137.
Santos, M., A. Ruiz, J. E. Quezada Díaz, A. Barbadilla and A. Fontdevila. 1992. The evolutionary history of Drosophila buzzatii. XX. Positive phenotypic covariance between field adult fitness components and body size. Journal of Evolutionary Biology 5:403-422.
Ruiz, A., M. Santos, A. Barbadilla, J. E. Quezada-Díaz, E. Hasson & A. Fontdevila. (1991). Genetic variance for body size in a natural population of Drosophila buzzatii, Genetics 128: 739-750.
Barbadilla, A. 1991. State of art of the problematic of higher taxa. Ameghiniana 28:295-301.
Barbadilla, A., J. E. Quezada Díaz, A. Ruiz, M. Santos and A. Fontdevila. 1991. The evolutionary history of Drosophila buzzatii. XVII. Double mating and sperm predominance. Genetics Selection Evolution 23:133 140.
Santos, M., A. Ruiz, A. Barbadilla, J. E. Quezada Díaz, E. Hasson and A. Fontdevila. 1988. The evolutionary history of Drosophila buzzatii. XIV. Larger flies mate more often in nature. Heredity 61:255 262.
Barbadilla, A. & H. Naveira. (1988). «The estimation of parental genotypes by the analysis of a fixed number of their offspring.», Genetics 119: 465-472.
Articles en llibres
Berry, A. & A. Barbadilla. (2000). «Gene conversion is a major determinant of genetic diversity at the DNA level», en Evolutionary Genetics from Molecules to Morphology. R. S. Singh y C. B. Krimbras (eds.) Cambridge University Press, NY
A. Barbadilla (2009). "Darwinismo y Creacionismo". Cent cinquanta anys després de L’origen de les espècies de Darwin, (A. Navarro y C. Segarra eds.). Treballs de la Societat Catalana de Biologia, 60 (2009) 245-253. Versión castellana aquí
A. Barbadilla (2008). "La recerca sobre el genoma humà". Enciclopèdia Catalana.
A. Barbadilla. (2003). "La Comunicación Social de la Ciencia e Internet" en La Ciencia es Cultura, págs. 177-180. Coordinador: Manuel Toharia, Edita: Soc. Gestión Museo de las Ciencias Príncipe Felipe de Valencia.
A. Barbadilla. (2000). "La selección natural: me replico, luego existo", en Evolución y filogenia de Artrópodos, pág. 605-612. SEA Vol. 26
A. Barbadilla. (1990). "La estructura de la Teoría de la Selección Natural", en Temas actuales de Biología Evolutiva, en A. Ruiz y M. Santos (Coordinadores), págs. 163-191. Publicaciones UAB, Barcelona.
Essays and popular articles
Barbadilla, A.; S. Casillas & A. Ruiz. 2019. La teoría neutralista de la evolución molecular, medio siglo después. Investigación y Ciencia. Págs. 52-59. Febrero 2019.
Web servers and genetic diversity databases
-
Murga-Moreno, J.; Coronado-Zamora, M.; Hervás, S.; Casillas, S & A. Barbadilla 2019. iMKT: the integrative McDonald and Kreitman test. URL: https://imkt.uab.cat/
-
Murga-Moreno, J.; Coronado-Zamora, M.; Bodelón, A.; Barbadilla, A. and Casillas, S. 2018. PopHumanScan: the online catalog of human genome adaptation. URL: http://pophumanscan.uab.cat
-
Casillas, S., R. Mulet, P. Villegas-Mirón, S. Hervás, E. Sanz, D. Velasco, J. Bertranpetit, H. Laayouni & A. Barbadilla. 2018. PopHuman: the human population genomics browser. URL: http://pophuman.uab.cat
-
PopFly: the Drosophila population genomics browser. S. Hervas, E. Sanz, S. Casillas, J. Pool and A. Barbadilla. 2017. URL: http://popfly.uab.cat
-
PopDrowser: The Population Drosophila Browser. Ràmia M, Librado P, Casillas S, Rozas, J & Barbadilla A. 2012. URL: http://popdrowser.uab.cat
-
Standard & Generalized McDonald and Kreitman test: a website to detect selection by comparing different classes of DNA sites. Egea, R., S. Casillas and A. Barbadilla. 2008. URL: http://mkt.uab.cat
- PDA: a pipeline to explore and estimate polymorphism in large DNA databases - Casillas, S. & A. Barbadilla. 2004. PDA: Pipeline Diversity Analysis. Casillas, S. & A. Barbadilla. 2006. PDA v.2: improving the exploration and estimation of nucleotide polymorphism in large data sets of heterogeneous DNA. URL: http://pda.uab.es
-
MamPol: Mammalia Polymorphism Database. Egea, R, S. Casillas, E. Fernández, MA Senar & A. Barbadilla (2006). URL: http://mampol.uab.es/
-
DPDB: Drosophila Polymorphism Database. URL: http://dpdb.uab.es / Casillas, S. & A. Barbadilla. 2005.
Elevator Pitch IBB-UAB 2019 by Marta Coronado
Funding
Additional information