A multilaminar model explains the structure of chromosomal aberrations
08/10/2015
During cell division, each metaphase chromosome contains a single enormously long DNA molecule that is associated with histone proteins and forms a long chromatin filament with many nucleosomes. Currentchromosome models consider that chromatin is folded forming loops or irregular networks. However, previous microscopy studies performed by researchers at the Chromatin Laboratory directed by professor Joan-Ramon Daban (Biosciences Unit of the Department of Biochemistry and Molecular Biology, UAB) showed unexpectedly that chromatin from metaphase chromosomes is folded into multilayered plates. This discovery led to the proposal of the thin-plate model, in which it is considered that chromosomes are formed by many stacked layers of chromatin oriented perpendicular to the chromosome axis. It was shown recently that this organization can justify the elongated cylindrical shape and mechanical properties of chromosomes.
Images obtained in many laboratories with different cytogenetic techniques have been used by Dr. Daban to investigate the internal structure of chromosomes. The orientation angle of G- and R-bands with respect to the chromosome axis has been measured; the mean values obtained for this angle range from 88o to 91o. This orthogonal orientation is observed even for the thinnest bands. The sub-bands produced by mechanical splitting of the original bands and the thin replication bands are also perpendicular to the chromosome axis. These results indicate that in three dimensions bands are disk-like structures that can be very thin and that short stretches of DNA can occupy completely the chromosome cross-section. Furthermore, in sister chromatid exchanges, the resulting connection surfaces are planar, and the observed orientation angle of these surfaces is approximately 90o. The connection surfaces observed in chromosome translocations occurring in different carcinomas and hematological malignancies are also planar; the mean values of the orientation angle of these surfaces in translocations analyzed using multicolor techniques (SKY and M-FISH) range from 90o to 92o.
All these observations impose geometric constraints that must be considered for the validation of models for chromatin organization in metaphase chromosomes. Models based on chromatin loops and irregular networks are not compatible with these constraints. The orthogonal orientation and planar geometry of the connection surfaces in chromosome translocations can be easily explained if chromosomes areformed by stacked thin layers of chromatin.This multilaminar model is also compatible with the orthogonal orientation of bands, with the existence of thin bands, and with band splitting produced by chromosome stretching.
This work has built a bridge between structural biology and cytogenetics. The results of this research are useful for the understanding of the three-dimensional organization of DNA in the chromosome and of the structural basis of the cytogenetic methods used for the diagnosis of hereditary diseases and cancers. The workhas been published in the multidisciplinary journal Scientific Reports from Nature Publishing Group.
Article:
Joan-Ramon Daban (2015) Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Scientific Reports 5:14891 www.nature.com/articles/srep14891.
Figure:
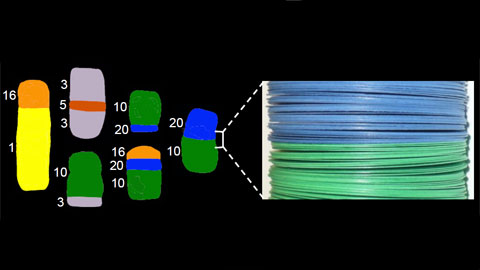
Direct relationship between the shape of translocations in cancer cells and the multilayer structure of chromosomes. The SKY images of different translocations involving chromosomes 1, 3, 5, 10, 16, and 20 in breast cancer cell line T47D were obtained from NCI-60 Drug Discovery Panel (http://home.ncifcrf.gov/CCR/60SKY/new/demo1.asp).