Discovery of a novel phage defence mechanism in Salmonella highlights emerging challenges for phage therapy
Phage therapy, the use of viruses that infect bacteria, is considered a promising strategy for the treatment of bacterial infections. However, it faces the challenge that bacteria can develop sophisticated defence mechanisms against phages. Researchers at the UAB have identified a new defence mechanism in Salmonella acquired through lateral gene transfer in the intestines of broiler chickens during oral phage therapy treatment.
Researchers from the Molecular Microbiology Group at the Department of Genetics and Microbiology of the UAB, led by Dr Montserrat Llagostera, have published a pioneering study describing a new defence mechanism acquired through lateral gene transfer in Salmonella, which leads a reduction in the expression of structural virion and lysis-associated proteins of UAB_Phi20 bacteriophage, resulting in abortive infection.
In recent years, phage therapy has become a key strategy for treating bacterial infections caused by pathogens that have developed multidrug resistance to conventional antimicrobials. However, the co-evolution of bacteriophages and bacteria has given rise to a broad array of bacterial strategies to counteract phage infection, which poses a significant challenge to the widespread application of phage therapy in both human and animal health. Studying these defence mechanisms is crucial for understanding phage–bacterium interactions and ensuring the safe use of phage therapy.
In this context, the recently published work represents an innovative study by identifying the ibfA gene as a novel defence factor in Salmonella enterica serovar Typhimurium against the virulent phage UAB_Phi20. This finding is particularly relevant because the gene is not encoded in the bacterial genome but has instead been identified in a conjugative plasmid belonging to the IncI1α group. This plasmid was acquired by Salmonella through lateral transfer in the intestine of broiler chickens during oral phage therapy treatment.
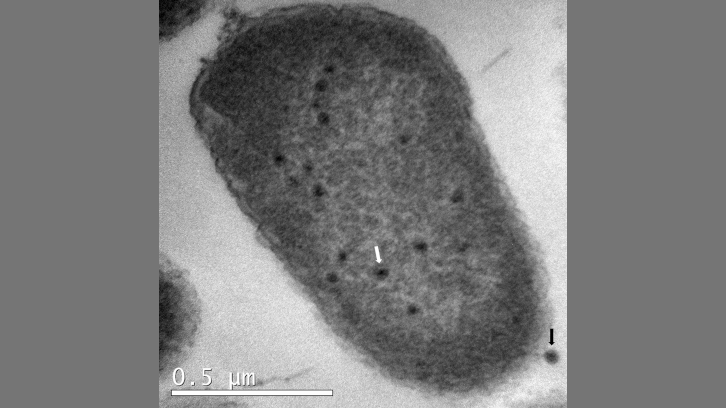
Using a methodological approach combining molecular microbiology, conventional microbiological techniques, transmission electron microscopy (TEM), and bioinformatics, the function of the ibfA gene has been characterised. This gene encodes a protein containing two domains: an ATPase-like domain and a TO-PRIM domain. Expression of IbfA significantly reduces the infectivity and productivity of phage UAB_Phi20, as evidenced by a reduction in efficiency of plating (EOP), efficiency of centre of infection (ECOI), and burst size, leading to decreased cellular viability without detectable cell lysis.
At the molecular level, it was observed that ibfA disrupts the transcription of the phage genome by promoting the expression of early phage genes such as the antirepressor ant. This causes an imbalance between the regulatory proteins Cro and C2, leading to a decrease in transcription of phage structural and cellular lysis proteins. Ultimately, this results in an abortive infection, an effect that could potentially extend to other P22-like phages.
Furthermore, although the evolutionary origin of ibfA remains uncertain, the gene is widely distributed across both chromosomes and plasmids of prokaryotes. This suggests that it may have additional beneficial functions for bacterial cells beyond its role in phage defence.
Departament of Genetics and Microbiology
Universitat Autònoma de Barcelona
References
López-Pérez, J.; Cortés, P.; Campoy, S.; Erill, I.; Llagostera, M. (2025). Deciphering the causes of ibfA-mediated abortive infection in the P22-like phage UAB_Phi20. International Journal of Molecular Sciences 26(10): 4918. https://doi.org/10.3390/ijms26104918

