Protein digestion grows up to assist in the study of the ubiquitinome
In our cells coexist thousands of proteins that act in a coordinated manner in adaptive processes in response to external or internal stimuli. This set of proteins is called proteome and the discipline that studies changes in the proteome to understand complex cellular processes is proteomics. Cells can modulate the activity of their proteins by regulating their levels (through the control of their synthesis or degradation) and also by post-translational modifications, which can determine changes in the activity of proteins, in their localization or in the interaction with other proteins.
There is a wide range of post-translational modifications, including the reversible addition of small chemical groups, as in the case of phosphorylation or acetylation, or small proteins such as ubiquitin. To interpret the role of post-translational modifications in cellular processes, it is necessary, on the one hand, to be able to detect experimentally which proteins are modified and, on the other hand, to determine how these modifications change under different experimental conditions.
In our research group we study the post-translational modifications involved in the activation processes of human T lymphocytes, a cellular model for the study of signal transduction and the immune response. Having characterized the phosphorylation events that take place upon T lymphocyte activation, the group initiated an ambitious project aimed at the characterization of other relevant post-translational modifications such as acetylation or ubiquitination (binding of ubiquitin to a substrate protein) and the study of the interaction between post-translational modifications. In this sense, it is known that ubiquitination plays an important role during T lymphocyte activation but the specific sites of modification and their dynamics have not been characterized on a large scale.
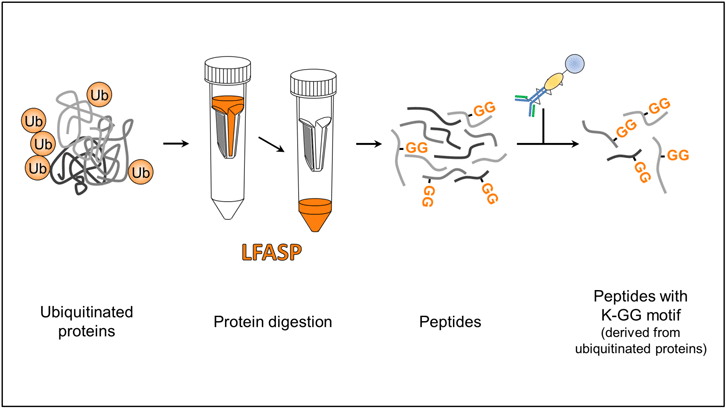
A widely used experimental strategy for the study of post-translational modifications consists in the enzymatic digestion of proteins into peptides (generally with trypsin), the subsequent purification of peptides containing a certain post-translational modification and finally the identification and localization of the modification using mass spectrometry. In the study of the ubiquitin-modified proteome (the ubiquitinome), trypsin digestion generates peptides containing the K-GG domain, in which two glycines (GG) derived from ubiquitin remain attached to the lysine (K) of the modified protein. This K-GG domain is recognized by specific antibodies that enable the purification of modified peptides and subsequent identification of ubiquitination sites in the protein. Thus, an efficient digestion is crucial in the generation of the K-GG motif recognized by the antibody.
There are different strategies for protein digestion, such as in-solution digestion or digestion in ultrafiltration units. Previous ubiquitinome studies have used in solution digestion and have started with large amounts of material (on the order of milligrams) to compensate the fact that modified peptides are low abundant and thus facilitate their detection. Protein digestion in ultrafiltration units (FASP) is a recent, more efficient method than in-solution protein digestion, but its application had been restricted to the digestion of small amounts of sample (on the order of micrograms).
We have developed a large-scale FASP method (LFASP) that allows digestion of sample on the order of milligrams and we have evaluated the applicability of this method to the study of the ubiquitinome. Using this methodology, we have identified about 12,000 ubiquitin peptides starting from 12 mg of protein extract with an average purification selectivity above 70%. Our results demonstrate that LFASP digestion is efficient, robust and reproducible. In addition, comparison with reference ubiquitinome studies demonstrates the superior digestion efficiency of the LFASP method.
We are currently using LFASP for the study of ubiquitinome dynamics during T lymphocyte activation. This research will reveal important clues about the molecular mechanisms involved in activation and signaling pathways in T lymphocytes and will open the door to the identification and characterization of new targets for the diagnosis and treatment of human diseases, including immunological diseases and cancer.
Acknowledgments:
This work was supported by Grant BIO2013-46492-R from the Spanish Ministry of Economy and Competitiveness. A.C. was the recipient of a Beatriu de Pinós Fellowship (Grant 2014 BP-B 00168) supported by the Universities and Research Secretariat of the Department of Economy and Knowledge of the Government of Catalonia and by the FP7 of the European Union through the Marie Curie Actions COFUND Programme. The Proteomics Laboratory CSIC/UAB is a member of ProteoRed, PRB2-ISCIII and is supported by Grant PT13/0001, of the PE I+D+i 2013-2016, funded by ISCIII and FEDER.
joaquim.abian.csic@uab.cat
Proteomics Laboratory CSIC/UAB
Institute of Biomedical Research of Barcelona (CSIC)
Universitat Autònoma de Barcelona
Albert Casanovas
albert.casanovas@iibb.csic.es
Proteomics Laboratory CSIC/UAB
Institute of Biomedical Research of Barcelona (CSIC)
References
Casanovas, Albert ; Pino-Llorente, Roberto; Carrascal, Monserrat; Abian Joaquin. Large-Scale Filter-Aided Sample Preparation Method for the Analysis of the Ubiquitinome. Anal. Chem., 2017, (89), (7), pp 3840–3846. DOI: 10.1021/acs.analchem.6b04804


