Described the potential activation mechanism of cannabinoid receptor 1
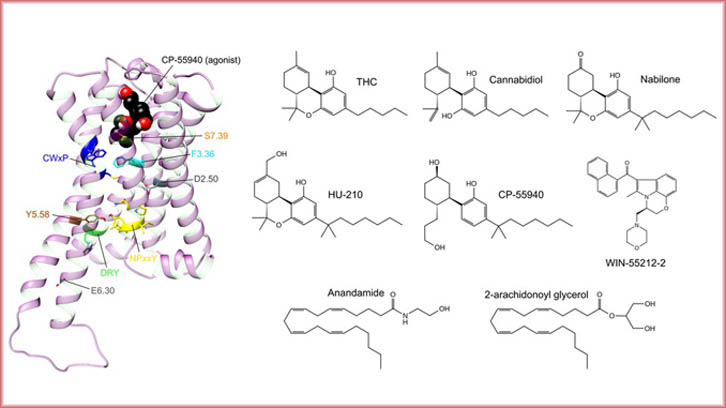
In these past years, there has been an increasing interest in the medical use of cannabis for the treatment of pain, nausea, vomit and neurological disorders. The main psychoactive compound of the plant, Δ9-tetrahydrocannabinol (THC), activates cannabinoid receptor 1 (CB1), one of the so-called G protein-coupled receptors (GPCRs). CB1 is mainly located at the central nervous system and is one of the most abundant receptors in the brain. Upon activation, a series of conformational changes occur at the level of receptor structure that allow binding and activation of a G protein, which in turn translates the activation message into a signaling cascade within the cell, leading to the eventual physiological response.
In physiological conditions, CB1 is regulated by molecules self-produced by the organism called endocannabinoids (such as anandamide and 2-arachidonoyl glycerol), but it can also be modulated by molecules found in cannabis plant extracts such as THC or cannabidiol (CBD). In order to find more potent and/or selective drugs, synthetic cannabinoids such as nabilone, HU-210, CP-55940 or WIN-55212 have been also developed. However, currently in the EU, only two cannabis-derived products (one in USA) and three synthetic products have been approved for medical use, which contain THC, CBD or nabilone as active ingredients.
One of the major disadvantages of novel drug discovery and development is its reliance on screening large collections of different molecules, resulting in lengthy, expensive and inefficient discovery programs. A possible solution to this problem is the use of structural knowledge of the drug target (in this case, the receptor), which allows to reduce the amount of tested molecules in preclinical trials. This entails obtaining the three-dimensional structure of the receptor (frequently by X-ray crystallography, nuclear magnetic resonance or electron microscopy methods), and/or predicting the putative binding of a molecule to the receptor by computational methods.
Currently, one of the most employed computational methods for studying protein function (including GPCRs) is molecular dynamics, which allows to observe changes in the three-dimensional structure of proteins at the atomic level and, therefore, understand their mechanisms of function.
This study describes the potential activation mechanism of CB1. Although some of the conformational changes that occur upon receptor activation had been previously identified, the complete transition from its inactive to its active state had not been observed until now. In this article, based on molecular dynamics simulations, the steps that take place from the binding of an activator molecule (agonist in pharmacological terms) to receptor activation are described. Understanding the receptor activation mechanism is key for developing drugs that modulate receptor function in a more efficacious and safer way.
Department of Pediatrics, Obstetrics and Gynecology and Preventive Medicine and Public Health
Universitat Autònoma de Barcelona
References
Díaz, Ó., Dalton, JAR and Giraldo, J. (2019). Revealing the Mechanism of Agonist-Mediated Cannabinoid Receptor 1 (CB1) Activation and Phospholipid-Mediated Allosteric Modulation. Journal of Medicinal Chemistry. 13;62(11):5638-5654. DOI: 10.1021/acs.jmedchem.9b00612.

