eMIRNA: a novel bioinformatic tool to detect microRNA genes
Pig farming is the sector with the greatest economic impact in Catalonia. For this reason, it is essential to deepen the understanding of the physiology of this animal. Thanks to a collaboration between the Center for Research in Agrigenomics (CRAG) and the UAB, a group of researchers has developed a new bioinformatics tool, eMIRNA, which makes it possible to locate and identify microRNA genes in pigs and other species. These genes are small molecules important in the regulation of numerous productive characters of interest. The tool also offers the opportunity to improve on the previously existing catalogs of these types of genes.
MicroRNAs are small RNA molecules whose main function is to regulate gene expression and adapt it to the physiological needs of tissues and cells. In this sense, microRNAs have the ability to bind to messenger RNAs promoting their degradation or repressing their translation to proteins in ribosomes. Furthermore, they can decisively influence the activation or repression of numerous metabolic pathways and regulate relevant and highly varied biological processes. It is therefore extremely important to identify and map the microRNAs genes present in the genome of a given species in order to characterize their sequence and biological function, as well as to detect mutations that could affect their regulatory activity.
One of the main obstacles that researchers face when studying microRNA genes is the lack of reliable catalogs in many species of huge productive interest, such as pigs, sheep or goats. This demonstrates the urgent need to develop computer tools to identify and characterize microRNA genes, from any animal genome, in order to build comprehensive catalogs which can be used for research purposes.
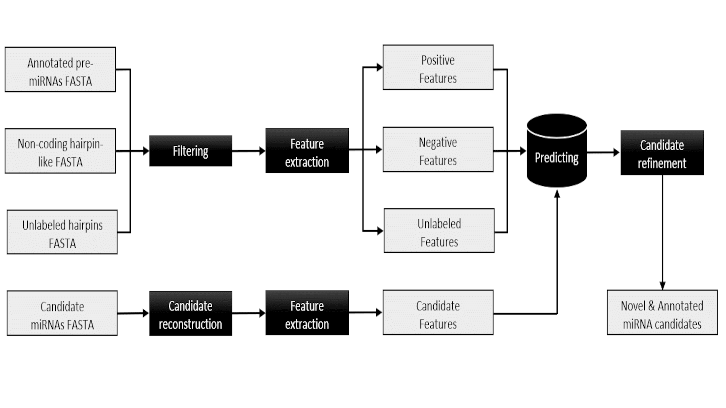
In this study, published in Genomics, we have developed a bioinformatics tool that allows the prediction and functional annotation of microRNAs. This tool includes different downloadable modules that are easy to use and adapt. A detailed description of this methodology can be found at: https://github.com/emarmolsanchez/eMIRNA. Our motivation stemmed from the need to delve into the annotation of microRNAs in the porcine genome, one of the domestic species with the greatest economic impact in the livestock sector in Catalonia.
One of the main innovations of the eMIRNA tool was to demonstrate the effectiveness of including a nucleotide motif search in the reconstruction of microRNA genes from massive sequencing and homology comparison data. Using a graph-based approach and a Machine Learning transduction algorithm, we were able to increase the amount of information available for the training of our classifier model, thus improving its predictive capacity compared to other previously published algorithms.
Furthermore, we demonstrate the practical applicability of this new tool using massive sequencing data from microRNAs in the skeletal muscle of Duroc pigs. A total of 47 microRNAs not included in the porcine catalog, and therefore completely unknown, were detected using the model implemented in eMIRNA, of which a total of 20 were identified in the porcine muscle samples.
Ultimately, the eMIRNA tool allows predicting with high reliability the microRNA genes of any animal species, as well as to functionally characterize their potential metabolic targets, thus providing the opportunity to improve the catalogs of microRNA genes in many species with still very limited genomic annotations. This task is essential to gain new insights, for instance, into the role of microRNAs in regulating numerous productive phenotypes of interest to the pig livestock sector.
Emilio Mármol Sánchez1 and Marcel Amills1,2
1Research Center of Agrigenomic.
2Universitat Autònoma de Barcelona.
References
Mármol-Sánchez E, Cirera S, Quintanilla R, Pla A, Amills M. (2020). Discovery and annotation of novel microRNAs in the porcine genome by using a semi-supervised transductive learning approach. Genomics 112: 2107-2118 doi: https://www.sciencedirect.com/science/article/pii/S0888754319304884?via%3Dihub


